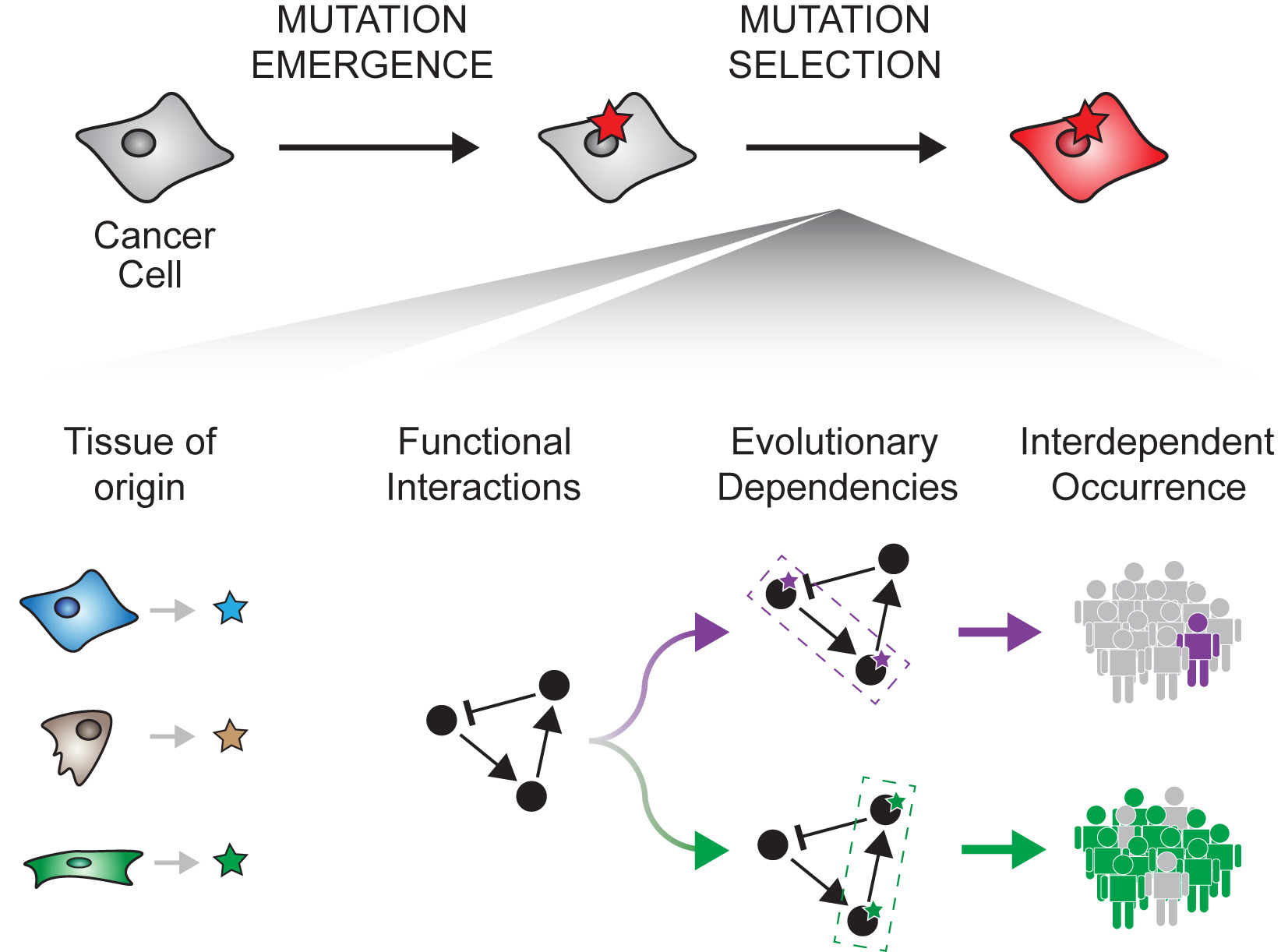
Cancer evolves through the emergence and selection of molecular alterations. Cancer genome profiling has revealed that specific events are more or less likely to be co-selected, suggesting that the selection of one event depends on the others. However, the nature of these evolutionary dependencies and their impact remain unclear. Previously, we designed SELECT (paper link), an algorithmic approach to systematically identify evolutionary dependencies from alteration patterns.
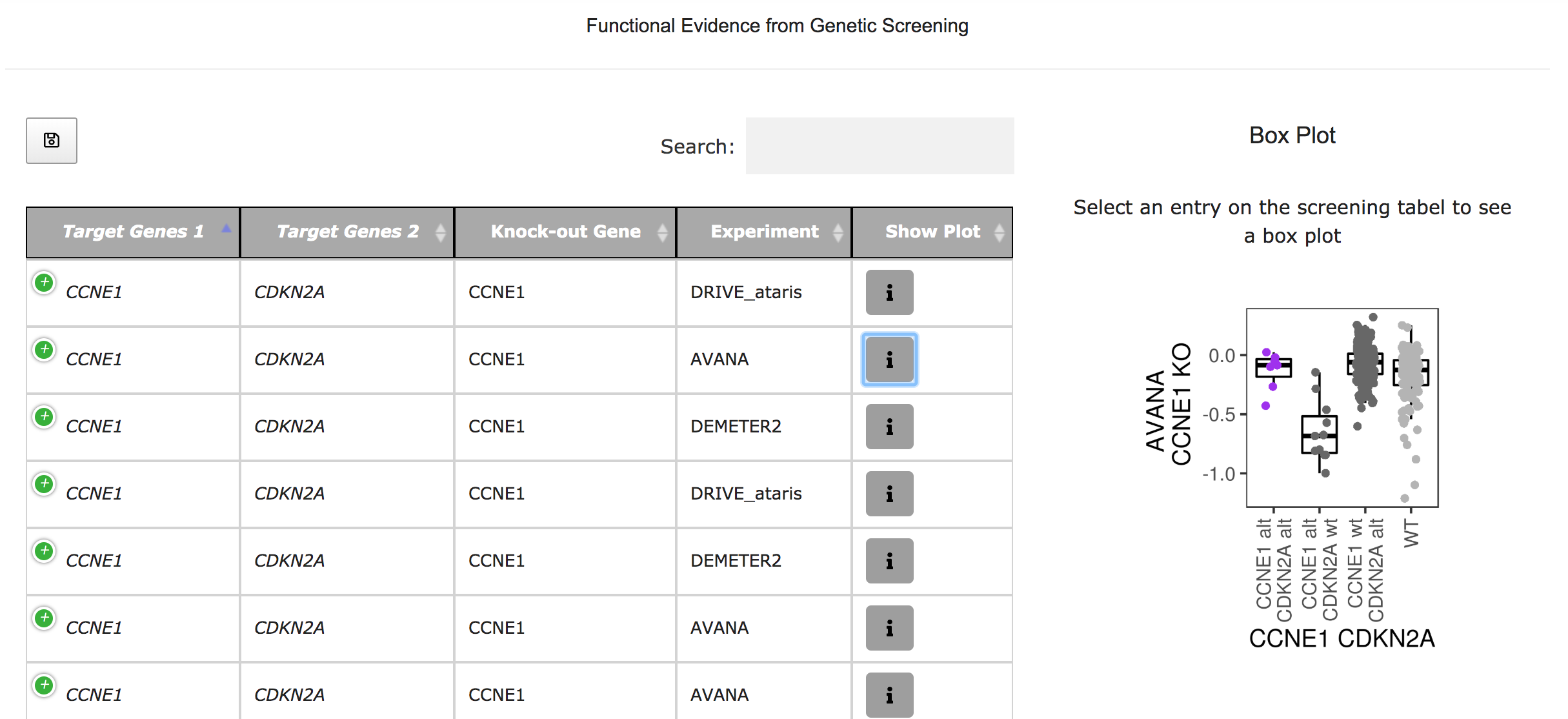
The systematic prediction and validation of advantageous alterations are major challenges in cancer genomics.In addition, most tumors harbor more than one oncogenic event. Hence it is crucial to understand which alterations act in concert during tumor progression, and functionally assess how their coexistence alters response to genetic and therapeutic perturbations. Here, we first integrated molecular profiles of >9,000 human cancer genomes to infer putative functional alterations and preferentially selected combinations of events. Next, using a Bayesian inference framework, we validated computational predictions with high-throughput functional readouts from genetic and pharmacological screenings on 2,000 cancer cell lines. Several significantly mutually exclusive and co-occurrent alterations in human tumors reflected, respectively, functional redundancies, able to rescue the phenotype of individual target inhibition, or synergistic interactions, which increased oncogene addiction. Among the top scoring dependencies, co-alteration of the PI3-kinase subunit PIK3CA and the nuclear factor NFE2L2 was a synergistic evolutionary trajectory in squamous cell carcinomas. In particular, double mutant cancer cell lines were associated with significantly different drug sensitivity profiles compared to cell lines with only one of the two alterations. By integrating computational, experimental, and clinical evidence, we provide a framework to study the combinatorial functional effects of cancer genomic alterations
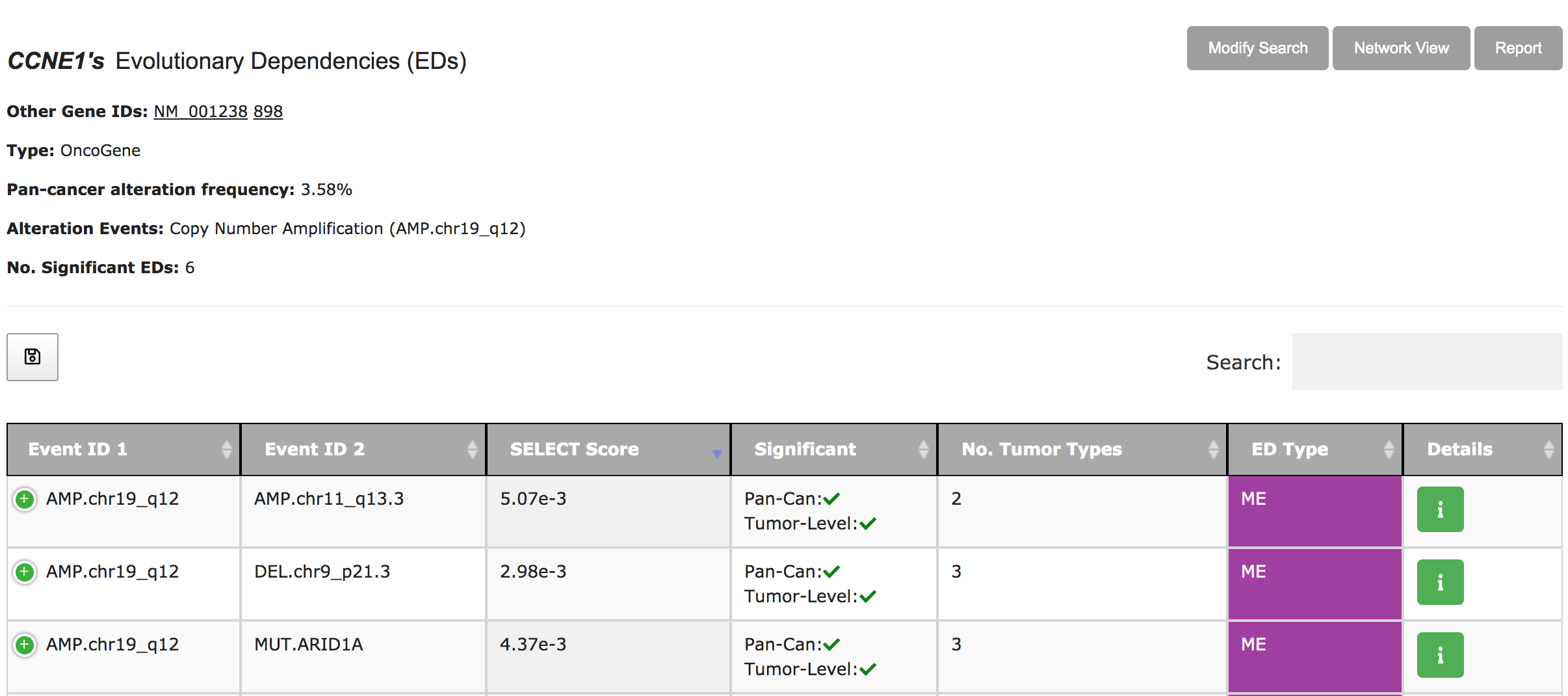
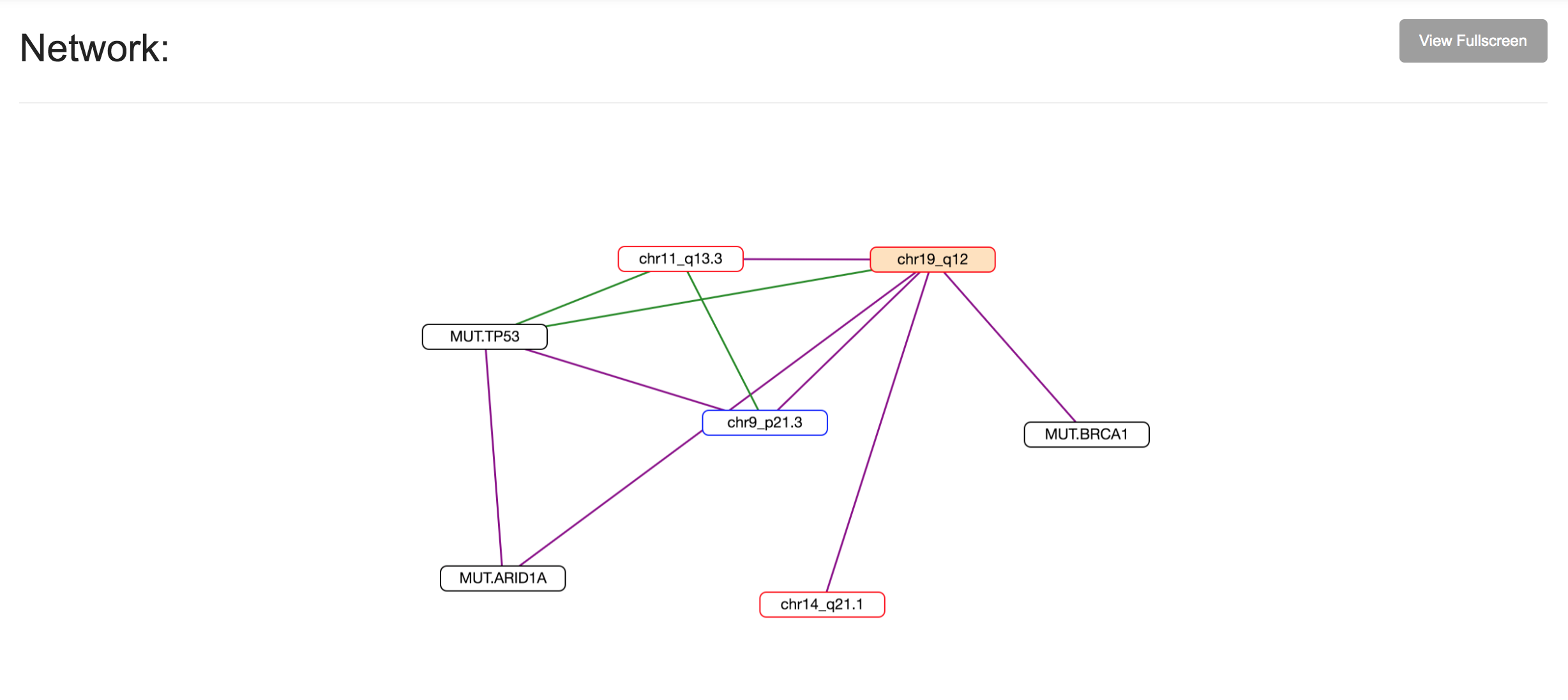
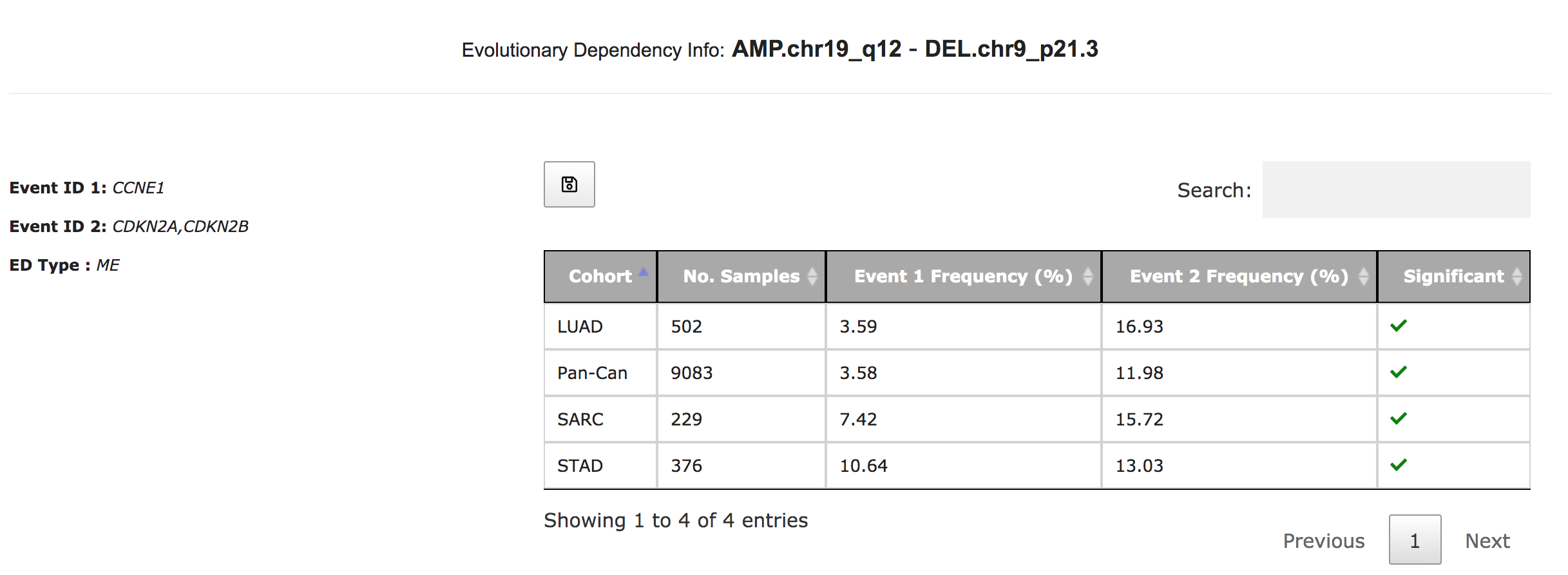
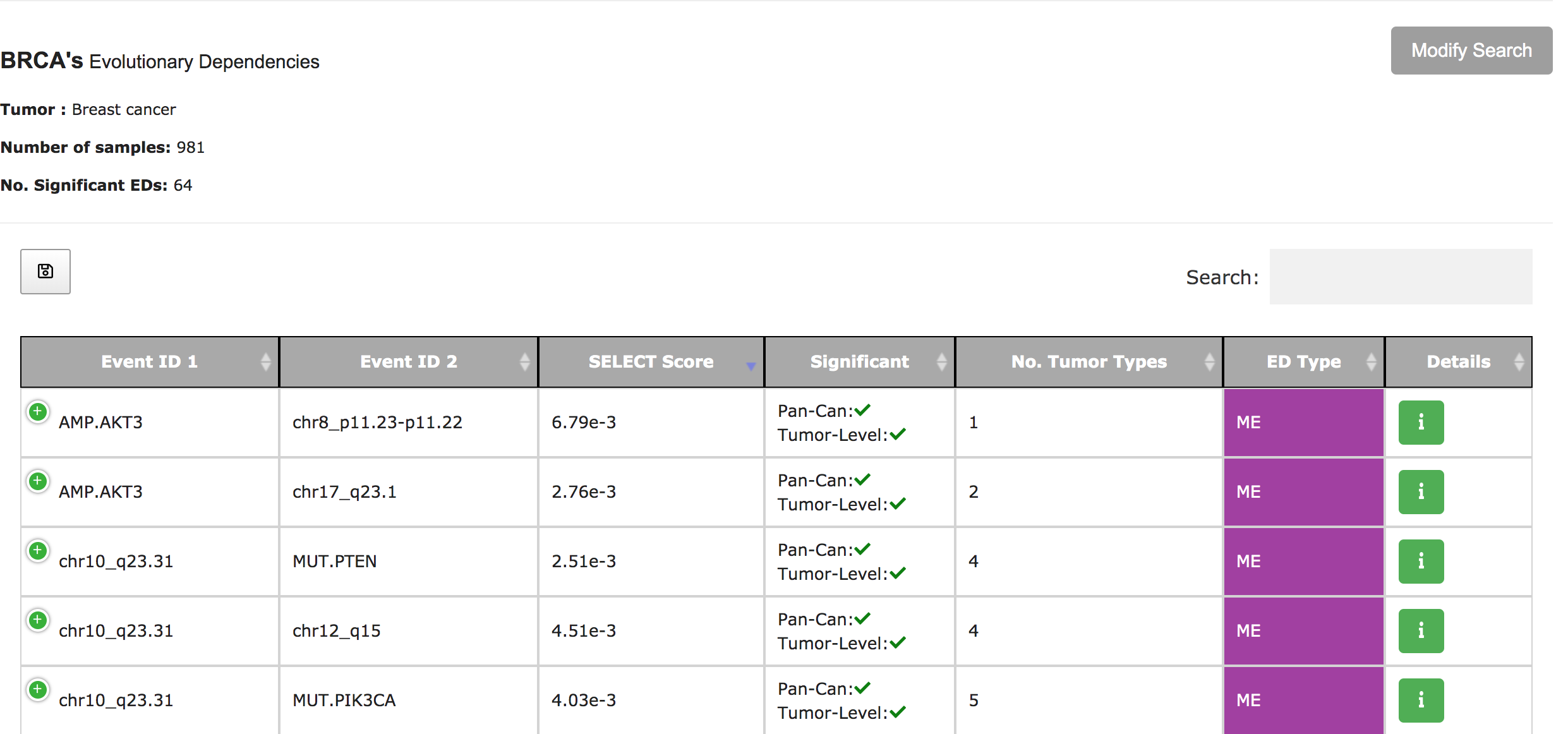
SELECT-Portal facilitates the exploration of EDs identified in the TCGA pan-cancer cohort, of which tumor types they are representative, and their effect on cell fitness and drug response